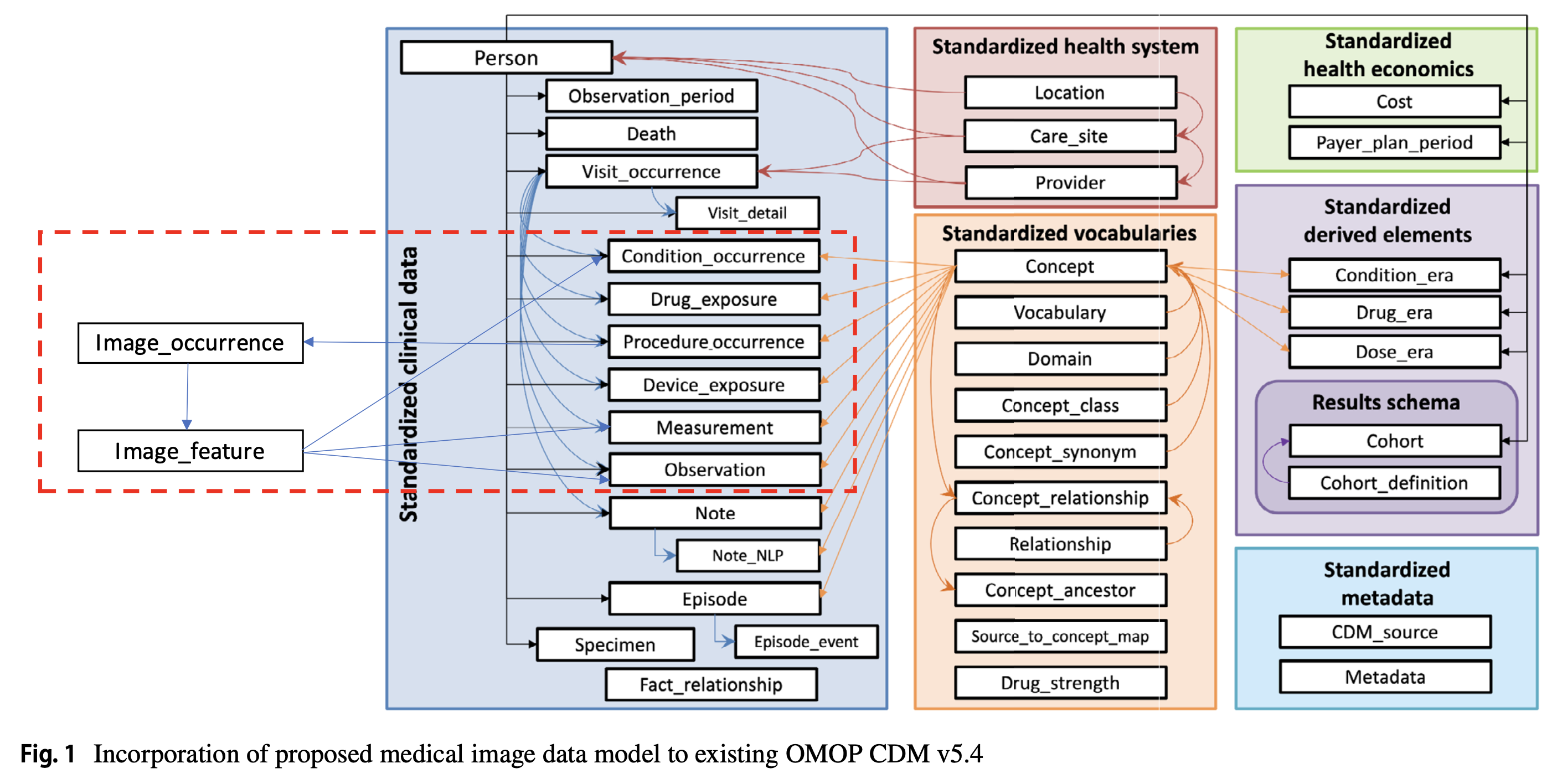
15 MI-CDM Basic
15.1 Person to Image feature
Q: Explaining how the link was established from Person → Visit_Occurence → Procedure_occurence → Image_occurence → Image_feature. Also, include the PK and FK of each table with example.
The path is patient-centric and flows like this:
Person → Visit_occurrence → Procedure_occurrence → Image_occurrence → Image_featureEach step uses primary keys (PK) and foreign keys (FK) to maintain referential integrity.
15.1.1 Person Table
Purpose: Identifies the patient.
Key fields:
person_id(PK) = unique identifier for each patient.
Example row:
| person_id | gender | birth_date |
|---|---|---|
| P001 | Male | 1965-03-12 |
15.1.2 Visit_occurrence Table
Purpose: Represents a healthcare encounter (e.g., outpatient visit, ER admission).
Keys:
visit_occurrence_id(PK)person_id(FK → Person)
Example row:
| visit_occurrence_id | person_id | visit_date | visit_type |
|---|---|---|---|
| V1001 | P001 | 2024-01-10 | Outpatient visit |
15.1.3 Procedure_occurrence Table
Purpose: Records that a procedure was performed (e.g., CT Chest).
Keys:
procedure_occurrence_id(PK)person_id(FK → Person)visit_occurrence_id(FK → Visit_occurrence)
Example row:
| procedure_occurrence_id | person_id | visit_occurrence_id | procedure_concept_id | procedure_date |
|---|---|---|---|---|
| PR5001 | P001 | V1001 | 36715236 (CT chest, SNOMED) | 2024-01-10 |
15.1.4 Image_occurrence Table (MI-CDM extension)
Purpose: Links procedure to the actual DICOM study/series.
Keys:
image_occurrence_id(PK)person_id(FK → Person)procedure_occurrence_id(FK → Procedure_occurrence)visit_occurrence_id(FK → Visit_occurrence)
Example row:
| image_occurrence_id | person_id | procedure_occurrence_id | visit_occurrence_id | study_UID | series_UID | modality_concept_id | anatomic_site_concept_id |
|---|---|---|---|---|---|---|---|
| IMG101 | P001 | PR5001 | V1001 | 1.2.840… | 1.3.6.1.4… | CT | Chest |
15.1.5 Image_feature Table (MI-CDM extension)
Purpose: Captures features/findings derived from the images.
Keys:
image_feature_id(PK)person_id(FK → Person)image_occurrence_id(FK → Image_occurrence)image_feature_event_id(FK → Measurement/Observation table)
Example row:
| image_feature_id | person_id | image_occurrence_id | image_feature_concept_id | image_finding_concept_id | value | alg_system | alg_datetime |
|---|---|---|---|---|---|---|---|
| F301 | P001 | IMG101 | LOINC: “Longest diameter of pulmonary nodule” | RadLex: “Pulmonary nodule” | 8 mm | AI-LungNodule-v1 | 2024-01-10 12:00 |
15.1.6 Putting It All Together
- Person: P001 is the patient.
- Visit_occurrence: P001 visits outpatient clinic on Jan 10, 2024.
- Procedure_occurrence: During visit, a CT Chest (SNOMED: 36715236) is ordered and performed.
- Image_occurrence: That procedure links to the actual DICOM study (UIDs, modality=CT, site=Chest).
- Image_feature: From that study, an AI algorithm extracts a pulmonary nodule feature (8 mm, solid).
👉 This chain ensures full traceability: We can follow the patient from clinical encounter → imaging procedure → DICOM study → extracted features.
15.2 Image feature to Clinical Table
Q: Show me how the Image_feature can be linked to the Condition_occurence, Measurement, and Observation table with example. Again, show PK/FK.
In the Image_feature table, two special fields are used for linkage:
image_feature_event_field_concept_id→ tells which clinical table the feature lives in (e.g., Measurement, Observation, Condition_occurrence).image_feature_event_id→ points to the primary key (PK) in that clinical table.
So, Image_feature acts like a bridge table that connects features to existing OMOP entities.
15.2.1 Link to Measurement Table
- Use case: Quantitative measurements from images (e.g., nodule diameter, CT attenuation).
Tables:
Measurement- PK =
measurement_id
- PK =
Image_feature- FK =
image_feature_event_id(points tomeasurement_id) - FK =
image_feature_event_field_concept_id(tells us this ID belongs to Measurement table).
- FK =
Example:
Measurement row:
measurement_id person_id measurement_concept_id value_as_number unit date M7001 P001 LOINC: “Longest diameter of pulmonary nodule” 8 mm 2024-01-10 Image_feature row:
image_feature_id image_occurrence_id image_feature_concept_id image_feature_event_id image_feature_event_field_concept_id F301 IMG101 LOINC: “Longest diameter of pulmonary nodule” M7001 Concept: Measurement
15.2.2 Link to Observation Table
- Use case: Qualitative or categorical descriptors (e.g., “solid vs ground-glass,” “spiculated margin”).
Tables:
Observation- PK =
observation_id
- PK =
Image_feature- FK =
image_feature_event_id(points toobservation_id)
- FK =
Example:
Observation row:
observation_id person_id observation_concept_id value_as_concept_id date O8001 P001 RadLex: “Pulmonary nodule margin” RadLex: “Spiculated” 2024-01-10 Image_feature row:
image_feature_id image_occurrence_id image_feature_concept_id image_feature_event_id image_feature_event_field_concept_id F302 IMG101 RadLex: “Spiculated” O8001 Concept: Observation
15.2.3 Link to Condition_occurrence Table
- Use case: Diagnoses inferred from imaging (e.g., “Lung cancer,” “Lung-RADS 4B”).
Tables:
Condition_occurrence- PK =
condition_occurrence_id
- PK =
Image_feature- FK =
image_feature_event_id(points tocondition_occurrence_id)
- FK =
Example:
Condition_occurrence row:
condition_occurrence_id person_id condition_concept_id condition_start_date C6001 P001 SNOMED: “Malignant neoplasm of upper lobe of lung” 2024-01-20 Image_feature row:
image_feature_id image_occurrence_id image_feature_concept_id image_feature_event_id image_feature_event_field_concept_id F303 IMG101 RadLex: “Lung-RADS 4B classification” C6001 Concept: Condition_occurrence
15.2.4 Big Picture
- Measurements: numeric imaging biomarkers (diameter, density, attenuation).
- Observations: categorical imaging descriptors (solid, spiculated, lobulated).
- Condition_occurrences: imaging-based diagnoses or classifications (lung cancer, Lung-RADS).
The Image_feature table ties them all together by recording which DICOM study/series they came from, what algorithm or report created them, and where in the OMOP model the actual values live.
15.3 Concept ID
Q: Explain what the concept ID (*_concept_id) is
Great question — concept IDs are central to how the OMOP CDM (and now the MI-CDM extension) work.
15.3.1 What is a *_concept_id?
- In OMOP CDM, every clinical concept (diagnosis, procedure, measurement, drug, imaging modality, anatomical site, etc.) is mapped to a unique integer identifier called a concept_id.
- These IDs live in the Concept table of OMOP.
- Instead of storing free-text or raw codes (like ICD-10, LOINC, SNOMED, DICOM tags, RadLex terms), OMOP standardizes them into a common vocabulary system.
15.3.2 Why use concept_id?
Standardization
- Different hospitals may record the same concept differently (ICD-10 vs SNOMED vs RadLex).
- OMOP maps them all to the same
concept_id-> ensuring interoperability.
Computable queries
- You can query across institutions without worrying about local coding differences.
- Example: “CT Chest” might be CPT 71250 in the US, but in Korea another code. Both map to the same OMOP
concept_id.
Relationships & Hierarchies
concept_idconnects toconcept_relationshipandconcept_ancestortables -> allowing you to query hierarchies (e.g., “find all imaging procedures that are CT scans of the chest”).
15.3.3 Where do you see *_concept_id?
In the MI-CDM tables:
15.3.3.1 Image_occurrence
modality_concept_id-> DICOM modality (e.g., CT, MR, US).anatomic_site_concept_id-> body part imaged (e.g., Chest, Liver).
15.3.3.2 Image_feature
image_feature_concept_id-> the specific feature (e.g., “Longest diameter of pulmonary nodule,” “Ground glass opacity”).image_feature_type_concept_id-> provenance (e.g., DICOM SR, AI algorithm, NLP extraction).image_finding_concept_id-> grouping (e.g., “Pulmonary nodule”).anatomic_site_concept_id-> location of the feature (e.g., “Left lower lobe”).
15.3.4 Example with Real IDs (simplified)
Suppose patient P001 has a CT chest with a lung nodule:
Procedure_occurrence
procedure_concept_id = 36715236-> “CT of chest”
Image_occurrence
modality_concept_id = 44814653-> “CT”anatomic_site_concept_id = 4144262-> “Chest”
Image_feature
image_feature_concept_id = 3026910-> “Longest diameter of pulmonary nodule (LOINC)”image_feature_type_concept_id = 45884084-> “Extracted by algorithm”image_finding_concept_id = 45756815-> “Pulmonary nodule (RadLex)”- Value = 8 mm
Key point: The concept_id makes sure that no matter the coding system (LOINC, SNOMED, RadLex, DICOM), the feature or event is normalized into a unified vocabulary for analytics.