Code
from pyhere import here
import nrrdThe NRRD (Nearly Raw Raster Data) file format is used to store and organize scientific and medical imaging data, commonly for 3D and 4D datasets. NRRD files are often used in applications like 3D Slicer for handling multi-dimensional imaging data. The format is flexible, supporting various data types, dimensions, and metadata.
.nrrd file or stored in a separate file, referred to by the header.An NRRD header typically looks like this:
NRRD0004
type: float
dimension: 3
sizes: 256 256 128
encoding: gzip
spacings: 1.0 1.0 1.5
axis-mappings: 0 1 2
kinds: space space space
space origin: (0,0,0)float, short, int).raw, gzip for compression).space for spatial dimensions).pynrrdfrom pyhere import here
import nrrdThe pynrrd Python library is a popular tool for reading, writing, and manipulating NRRD files. Below are the steps to install pynrrd and use it to read NRRD files.
To read an NRRD file, use the nrrd.read() function, which returns two items:
Here’s a basic example:
# Read the NRRD file
data, header = nrrd.read(here("data/MRI-Brain-Eye/Case1/3 t1 axial Processed_CaPTk.nrrd"))
# Display the shape of the data
print("Data shape:", data.shape)
# Display the header information
print("Header metadata:", header)Data shape: (192, 256, 192)
Header metadata: OrderedDict([('type', 'unsigned short'), ('dimension', 3), ('space', 'left-posterior-superior'), ('sizes', array([192, 256, 192])), ('space directions', array([[ 0.97656101, -0.00170442, 0. ],
[ 0.00170442, 0.97656101, 0. ],
[ 0. , 0. , 1. ]])), ('kinds', ['domain', 'domain', 'domain']), ('endian', 'little'), ('encoding', 'gzip'), ('space origin', array([ -89.87082095, -145.95727884, -78.06524467]))])data: This will be a multi-dimensional NumPy array, with dimensions specified by header['sizes']. You can directly manipulate this array or use it for visualization.
header: A dictionary containing the metadata extracted from the NRRD header, including keys like type, dimension, spacings, and space origin.
You can access specific metadata fields from the header dictionary:
# Access voxel spacing
spacing = header.get('spacings', None)
print("Voxel Spacing:", spacing)
# Access data type
data_type = header.get('type', None)
print("Data Type:", data_type)Voxel Spacing: None
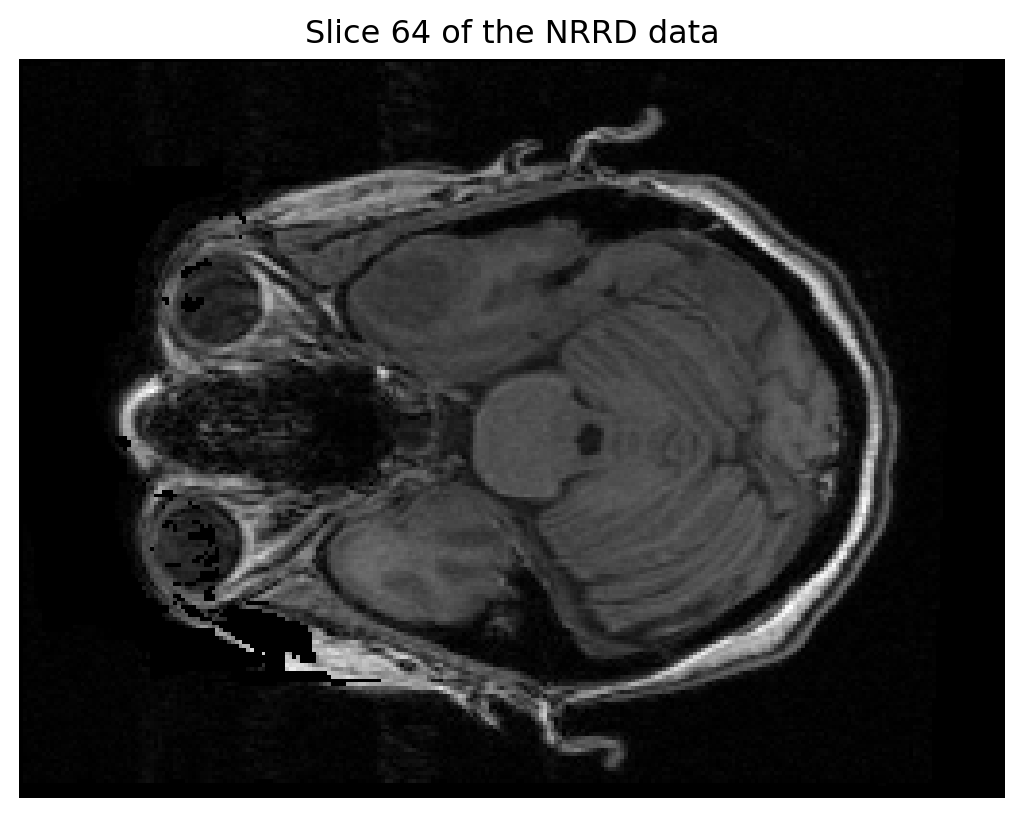
Data Type: unsigned shortTo visualize a slice of the 3D NRRD data (for example, slice 64 along the z-axis), you can use matplotlib:
import matplotlib.pyplot as plt
# Select a slice from the 3D data
slice_data = data[:, :, 64]
# Display the slice
plt.imshow(slice_data, cmap='gray')
plt.title("Slice 64 of the NRRD data")
plt.axis('off')
plt.show()